Data source
The Japan Cardiovascular Database-Keio Interhospital Cardiovascular Studies (JCD-KiCS) is a large, ongoing, prospective multicenter (n = 15) PCI registry to collect clinical data of consecutive patients undergoing PCI in Japan that developed in collaboration with the National Cardiovascular Data Registry (NCDR) CathPCI9,10,11. In JCD-KiCS, all PCI procedures were conducted under the direction of the intervention team of each participating hospital according to standard care. Participating hospitals were instructed to register data from consecutive PCI using an electronic data-capturing software system equipped with a data query engine and validations to maintain data quality. Data entry was conducted by dedicated clinical research coordinators who trained for JCD-KiCS specifically. Data quality was ensured through the use of an automatic validation system and bimonthly standardized education and training for the clinical research coordinators. The senior study coordinator (I.U.) and extensive on-site auditing by the investigator (S.K.) ensured proper registration of each patient. The protocol of this study was under the principles of the Declaration of Helsinki and approved by the Keio University School of Medicine Ethics Committee and the committee of each participating hospital (National Hospital Organization Review Board for Clinical Trials; the Eiju General Hospital Ethics Committee; the Ethics Committee of Saiseikai Utsunomiya Hospital; the Research Ethics Committee, Tokyo Saiseikai Central Hospital; the Japanese Red Cross Ashikaga Hospital Ethics Committee; Kawasaki Municipal Hospital Institutional Review Board; Saitama City Hospital Ethical Review Board; Isehara Kyodo Hospital Institutional Review Board; Tokyo Dental College Ichikawa General Hospital Institutional Review Board; the Independent Ethics Committee of Hiratsuka City Hospital; The Saint Luke’s Health System Institutional Review Board; the Hino Municipal Hospital Institutional Review Board; and the Ethics Committee of Yokohama Municipal Citizen’s Hospital). All participants were provided verbal or written consent for the baseline data collection, and informed consent was obtained from all participants individually.
Study population
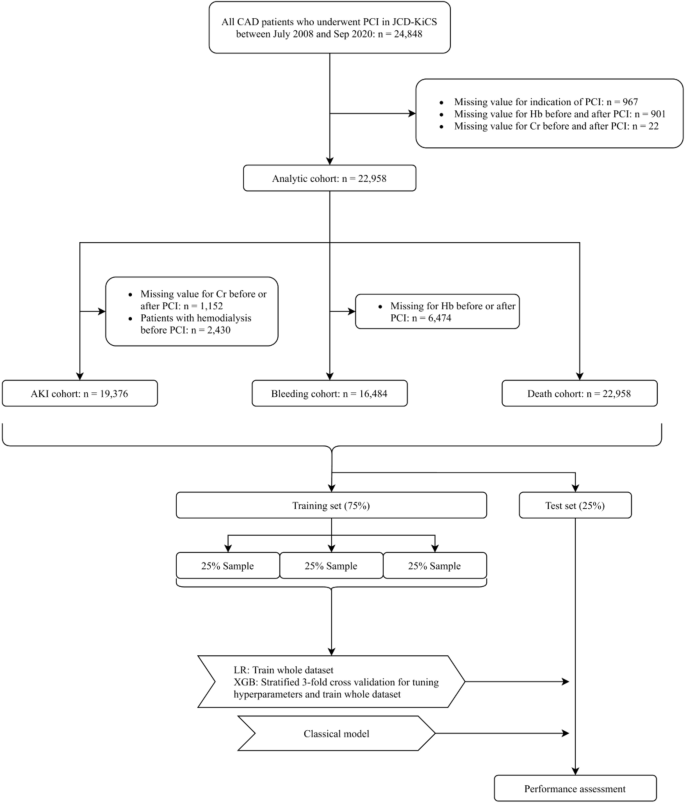
We extracted 24,848 consecutive patients who underwent PCI between July 2008 and September 2020. Because several parameters are applied as input variables for one model and the exclusion criteria of other models (e.g., hemodialysis before PCI is an input variable of the in-hospital mortality model and exclusion criteria of the AKI model), we made each outcome-specific cohort using a two-step procedure. First, we excluded patients with missing indications (n = 967), those without pre- and post-procedure hemoglobin (n = 901), and those without pre- and post-procedure serum creatinine (n = 22) (analytic cohort). Next, we applied outcome-specific exclusion criteria, followed by the imputation of missing values to make each cohort (detailed in Fig. 1). Each population was randomly split into a training set of 75% of the patients and a test set of the remaining 25% of the patients with approximately the same proportion of events.
Study flowchart. Abbreviations: CAD, coronary artery disease; PCI, percutaneous coronary intervention; JCD-KiCS, The Japan Cardiovascular Database-Keio Interhospital Cardiovascular Studies; Hb, aemoglobin; Cr, creatinine; AKI, acute kidney injury; LR logistic regression model; XGB, extreme gradient boosting model.
Definitions and outcomes
The definition of AKI, bleeding, and in-hospital mortality were consistent with original NCDR-CathPCI models4,5,6. Briefly, AKI was defined as a ≥ 0.3 mg/dl absolute or as a ≥ 1.5-fold relative increase in post-PCI creatinine or new dialysis initiation. Bleeding was defined as any of the following occurring within 72 h after PCI or before hospital discharge (whichever occurs first): site-reported arterial access site bleeding; retroperitoneal, gastrointestinal, genitourinary bleeding, intracranial hemorrhage, cardiac tamponade, or post-procedure hemoglobin decrease of 3 g/dl in patients with pre-procedure haemoglobin ≤ 16 g/dl, or post-procedure non-bypass surgery-related blood transfusion for patients with a pre-procedure haemoglobin ≥ 8 g/dl. In-hospital mortality was defined as any post-procedural death at the same hospital admission. Because JCD-KiCS was developed in collaboration with NCDR-Cath PCI, the majority of clinical variables were defined in accord with the data dictionary (version 4.1)9. For example, cardiogenic shock was defined as a sustained (> 30 min) episode of systolic blood pressure of < 90 mm Hg, and/or cardiac index of < 2.2 L/min/m2 determined to be secondary to cardiac dysfunction, and/or the requirement for intravenous inotropic or vasopressor agents or mechanical support to maintain the blood pressure and cardiac index above the specified levels within 24 h after the procedure.
Handling missing data
After enrollment of the analytic cohort, we imputed the missing value of pre-procedural hemoglobin with the value of post-procedural hemoglobin for the developed AKI and in-hospital mortality model, and imputed missing values of pre-procedural creatinine with those of post-procedural creatinine for the developed bleeding and in-hospital mortality models. Given that the absence rate was < 5% for any other variables, we handled the missing values to use a median imputation for the continuous variables and mode imputation for the categorical variables.
Model development
We developed two models: LR models and extreme gradient descent boosting (XGB) models. XGB is an ML algorithm that creates a series of relatively simple decision trees combined with boosting methods to develop more robust final predictions. In the LR model, we used the same categorized variables of the original NCDR-CathPCI risk scores (original model), and in the XGB model, we used the same variables but treated raw continuous variables that were categorized in the original models. The full list of variables was as follows:
-
1.
AKI model: age (categorized as < 50, 50–59, 60–69, 70–79, 80–89, and ≥ 90 years), heart failure within 2 weeks, estimated glomerular filtration rate (eGFR) (categorized as < 30, 30–44, 45–59, and ≥ 60 ml/min/1.73 m2), diabetes mellitus, prior heart failure, prior cerebrovascular disease, non ST-elevation acute coronary syndrome (NSTEACS), ST-elevation myocardial infarction (STEMI), cardiogenic shock at presentation, cardiopulmonary arrest at presentation, anemia defined as hemoglobin at admission of less than 10 g/dL, and use of IABP.
-
2.
Bleeding model: STEMI, age (categorized as < 60, 60–70, 71–79, and ≥ 80 years), BMI (categorized as < 20, 20–30, 30–39, and ≥ 40 kg/m2), prior PCI, eGFR (categorized as < 30, 30–44, 45–59, and ≥ 60 ml/min/1.73 m2), cardiogenic shock at presentation, female sex, hemoglobin at presentation (categorized as < 13, 13–15, ≥ 15 g/dL), and PCI status (Emergency, Salvage, Urgency, and Elective).
-
3.
In-hospital mortality model: age (categorized as < 60, 60–69, 70–79, and ≥ 80 years), cardiogenic shock at presentation, prior heart failure, peripheral artery disease, chronic obstructive pulmonary disease, estimated GFR (categorized as < 30, 30–44, 45–59, 60–89, and ≥ 90 ml/min/1.73 m2), NYHA classification IV at presentation, STEMI, and PCI status (emergency, salvage, urgency, and elective).
To optimize the hyperparameters of the XGB model, we used a stratified threefold cross-validation with a random search. After determining the best hyperparameters, XGB models were developed using the entire training set (hold-out methods, Supplementary Material for a more detailed explanation). In addition, we constructed the expanded LR and XGB models using additional variables selected by clinical significance. The additional variables were as follows:
-
Expanded AKI model: contrast volume and timing of PCI (i.e., during working or holiday times).
-
Expanded bleeding model: number of antiplatelet agents, use of anticoagulants at PCI, and timing of PCI.
-
Expanded in-hospital mortality model: technical failure of PCI, defined as failure to cross the guidewires or when the TIMI grade after PCI was 1 or 0 (slow flow or no flow), and the timing of PCI.
Statistics and key metrics
Continuous variables were summarized as medians with interquartile ranges and compared using Mann–Whitney U tests, and categorical variables were summarized as frequencies and compared using chi-square tests or Fisher’s exact tests, as appropriate.
The C-statistics with 95% confidence intervals (95%CIs) based on the Delong method and the area under the precision-recall area under curve (PRAUC) were used to estimate the model discrimination. Model calibration was assessed using the Brier score and calibration plot. The Brier score is defined as the mean squared difference between the observed and predicted outcomes and ranges from 0 to 1.00, with 0 representing the best possible calibration. The two primary components decomposed from the Brier score, i.e., reliability and resolution, were also evaluated. Calibration plots were used to plot the mean risk score relative to the observed outcome rate for a given quintile of the predicted risk. Furthermore, we used the net reclassification index (NRI) to evaluate the clinical utility of the LR and XGB models with cut-off values of 10%, 4%, and 2.5% for AKI, bleeding, and in-hospital mortality, respectively. A P value of < 0.05 was considered statistically significant. This study is based on the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) guidelines.
Sensitivity analysis
We used a multiple imputation method to handle missing values instead of a median imputation method. The multiple imputation model included all prespecified predictors and outcomes as recommended12. Ten imputed datasets were generated, and the C-statistics were combined using Rubin’s rules.
Software Implementation
All analyses were conducted in R (version 4.0.4; R Project for Statistical Computing, Vienna, Austria) with tidymodels (version 0.1.2) bundle of packages for data pre-processing, hyperparameter tuning, learning, and performance metrics13,14,15. We used xgboost (version 1.3.2.1) for extreme gradient descent boosting16, pROC (version 1.17.0.1) to calculating C-statistics17, verification (version 1.42) to calculate Brier scores18, predictABEL (version 1.2.4) to calculate the NRI19 mice (version 3.14.0) to perform multiple imputation20.